This post demostrate implements the computation and visualization of CSI (Contamination Severity Index) develop by Dr. P.K Sen in 2016, data obtain from British Geological Survey.
Map getting from GADM version 2.8
load('data.Rda')
thana_map <- readRDS(gzcon(url('https://biogeo.ucdavis.edu/data/gadm2.8/rds/BGD_adm3.rds')))1. Expression
Let \(Y_{(i)}, i = 1,2, \ldots, n\) be the ordered sample from a region (can be division/district/thana), in our case, thana.
- Let \(M\) denote the number of observation less than the threshold \(L\) (given).
- Let \(F_n(x) = \frac{1}{n}\sum_{i = 1}^{n}I(Y_i \leq x), x \geq 0\), the empirical distribution be the estimator of true distribution \(F(x)\).
For arsenic (As) level \(Y_{(i)}\), the corresponding propensity socre, denoted by \(C_{ni}, i = 1,2, \ldots,n\) is given by
\[\begin{align} \tag{2.1} C_{ni} = \max(0, (1 - L/Y_i)^{\hat{\theta(Y_{(i)})}}) \end{align}\]
where
\[\begin{align} \tag{2.2} \hat{\theta(Y_{(i)})} = \frac{2}{n-M}\sum_{j = M+1}^{n}\frac{\min(Y_{(i)},Y_{(j)})}{Y_{(i)}+ Y_{(j)}}, i = 1,\ldots, n \end{align}\]
and finally the CSI is the mean of the propensity score averaged by all \(n\) observation.
\[\begin{align} \tag{2.3} \hat{C}_E = \frac{1}{n}\sum_{M \leq i \leq n}C_{ni} \end{align}\]
Note: For Equation (1), if \(Y_{(i)} \leq L\), then \(C_{ni} = 0\).
2. Implementation
propensity_score <-function(x, y_vector, L = 10){
# x is argument, i.e. oberservation to be convert
# y_vector is a vector of ALL observations in the sub_region
n <- length(y_vector)
y_truncate <- y_vector[y_vector > L]
# case that ALL observation less than the threshold
if(length(y_truncate) == 0)
return(rep(0, n))
# when num of y_trancate == 1, then 2/(n-M-1) = infinity
if(length(y_truncate) == 1)
return(max(0,1-L/x))
# base of Eq. (1)
C_ni <- max(0,1-L/x)
# coeff * y_sum is Eq. (2)
coeff <- 2/length(y_truncate)
y_sum <- sum(pmin(x,y_truncate)/(x + y_truncate))
# return value of Eq. (1)
return(C_ni^(coeff*y_sum))
}Once we have the CSI score for the thana, CSI is simply the mean of the scores of all observations.
csi <- function(y_vector){
# find propensity score for each observation
score <- sapply(y_vector, y_vector = y_vector, propensity_score)
# mean of the score is csi
return(mean(score))
}3. Result
We present two type of plot,
- The first one we use the exact CSI values value computed from the estimator, it is presented in a continuous scale.
library(tibble)
library(ggplot2)
library(dplyr)
thana_obs <- split(dataset, dataset$thana_id)
thana_csi <- function(data_list, discrete = FALSE){
n <- length(data_list)
csi_vector <- rep(NA, n)
ids <- names(data_list)
for(i in 1: n){
csi_vector[i] <- csi(thana_obs[[i]]$as)
}
cutoff <- seq(0, 1, length.out = 11)
if (discrete == TRUE){
csi_vector <- cut(csi_vector, breaks = cutoff, include.lowest = TRUE)
csi_vector <- factor(csi_vector, levels = rev(levels(csi_vector)))
}
# can consider rounding to 3 decimals
df <- tibble(id = ids, csi = csi_vector)
return(df)
}
# csi in dataframe
csi_df <- thana_csi(thana_obs)
# plotting result
thana_df <- fortify(thana_map)
# merge result
plot_csi <- left_join(thana_df, csi_df, by = 'id')
dcsi_df <- thana_csi(thana_obs, discrete = TRUE)
plot_dcsi <- left_join(thana_df, dcsi_df, by = 'id')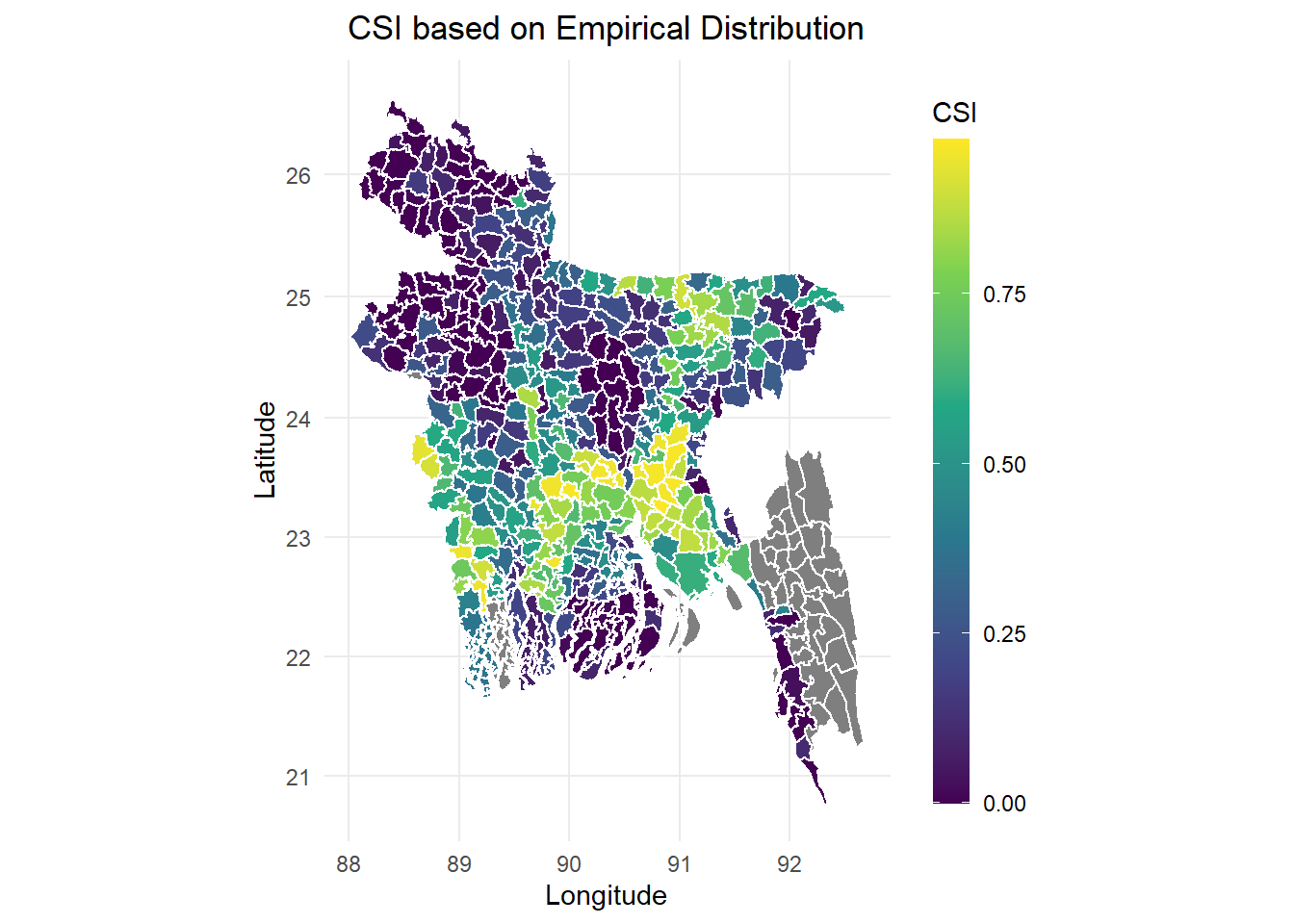
- The second one we use the exact 10 bins, from which we divide 0 - 1 into 10 categories, it is presented in a discrete scale.
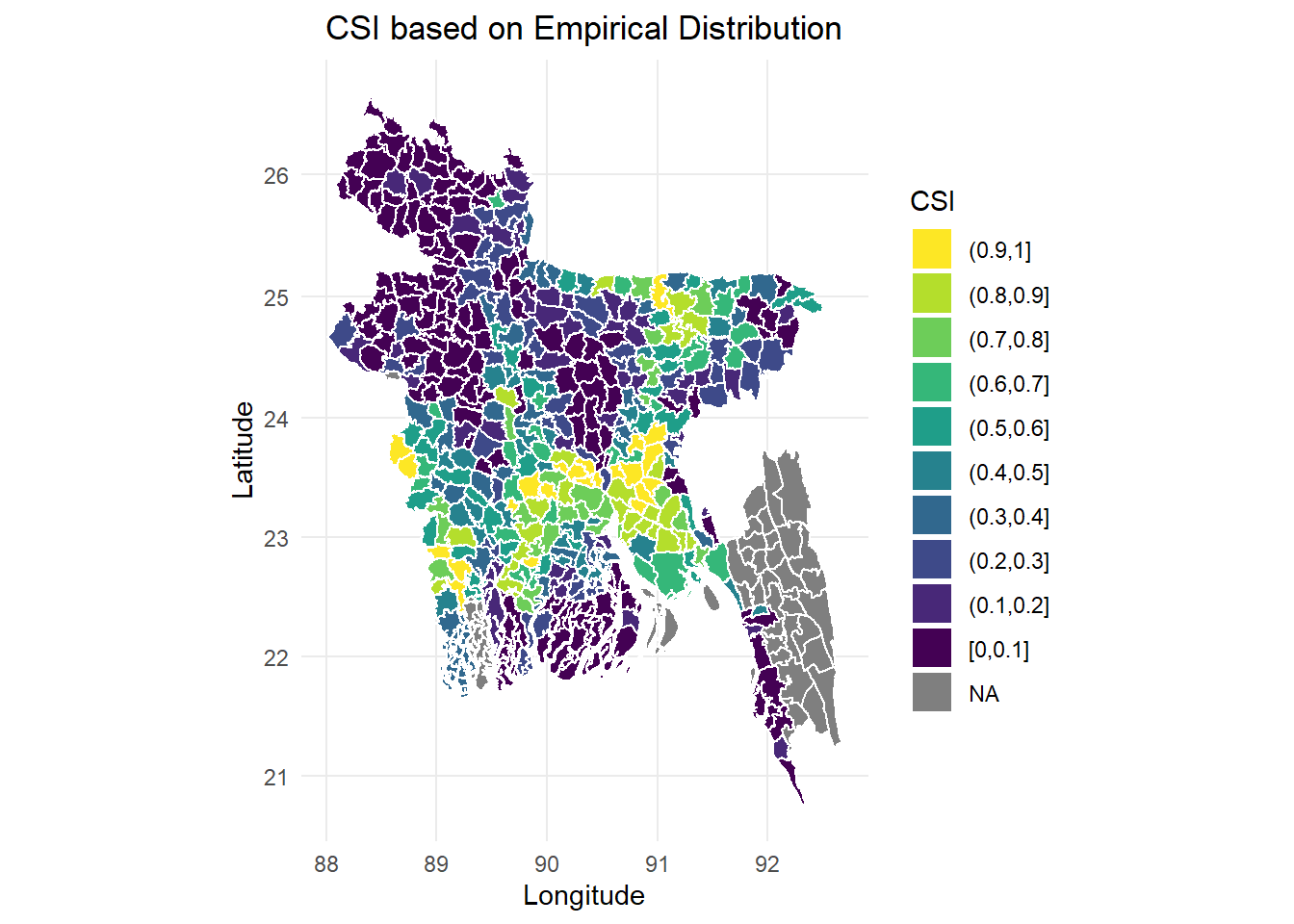
References
Sen, P. K. (2016). Abundant Environmental Arsenic Contamination: Some Statistical Perspectives. Sankhya B, 78(2), 341-361.