This post is a demonstration of fitting a Linear Mixed model (LMM) under Bayesian paradiagm. Linear mixed models are an extension of simple linear models to allow both fixed and random effects, and are particularly used when there is non independence in the data, such as longitudinal data.
1. Prelimineray
Formulation
In simple linear model of \(n\) observations, we have
\[\begin{align} \boldsymbol{y}&= \boldsymbol{X}\boldsymbol{\beta}+ \boldsymbol{\varepsilon}\\ \boldsymbol{\varepsilon}& \sim N(\boldsymbol{0}, \sigma^2 \boldsymbol{I}) \end{align}\]
where
\(\boldsymbol{y}= (y_1, \ldots, y_n)'\) is the response vector of \(n \times 1\);
\(\boldsymbol{X}\) is the model matrix, with typical row \(\boldsymbol{x}_i = (x_{1i}, \ldots, x_{pi})\) of \(n \times p\) ;
\(\boldsymbol{\beta}= (\beta_1, \ldots, \beta_p)\) is the vector of regression coefficients of \(p \times 1\);
\(\boldsymbol{\varepsilon}= (\varepsilon_1, \ldots, \varepsilon_n)\) is the vector of errors of \(n \times 1\);
\(\boldsymbol{0}\) is zero vector and \(\boldsymbol{I}\) is the identity matrix dimension \(n\).
In linear mixed models, we relaxes the constrain of independence and allow the so-called ``random effects’’ among subjects, and observations is a mixed of fixed and random effect. Hence the model becomes
\[\begin{align} \boldsymbol{y}_i &= \boldsymbol{X}_i\boldsymbol{\beta}+ \boldsymbol{Z}_i\boldsymbol{b}_i + \boldsymbol{\varepsilon}_i \\ \boldsymbol{b}_i &\sim N(\boldsymbol{0}, \boldsymbol{\Psi})\\ \boldsymbol{\varepsilon}_i & \sim N(\boldsymbol{0}, \sigma^2 \boldsymbol{\Lambda}_i) \end{align}\]
where \(n_i\) is the number of observation in subject (group) \(i\) and \(J\) is the number of distinct subjects,
\(\boldsymbol{y}_i\) is the \(n_i \times 1\) response vector for observations in the \(i\) th subject.
\(\boldsymbol{X}_i\) is the \(n_i \times p\) model matrix for the fixed effects for observations in subject \(i\).
\(\beta\) is the \(p \times 1\) vector of fixed-effect coefficients.
\(\boldsymbol{Z}_i\) is the \(n_i \times q\) model matrix for the random effects for observations in subject \(i\).
\(b_i\) is the \(J \times 1\) vector of random-effect coefficients for subject \(i\).
\(\boldsymbol{\varepsilon}_i\) is the \(n_i × 1\) vector of errors for observations in subject \(i\).
\(\boldsymbol{\Psi}\) is the \(J \times J\) covariance matrix for the random effects.
\(\sigma^2\boldsymbol{\Lambda}_i\) is the \(n_i \times n_i\) covariance matrix for the errors in subject \(i\).
In general, there are three common ways to estimate parameters,
Likelihood method, which may involved Restricted Maximum Likelihood for estimating the variance components.
lmerpackage is used commonly.Generalized Estimating Equation which does NOT assume distribution of the response,
geepackis prefered in general.Under Bayesian paradigm, it is easier to see LMM as a hierachical model structure, suppose for a varying-intercept, varying-slope model can be written as
\[\begin{align} Y_{ij} &\sim N(\alpha_j + \beta_{j} \cdot x_{ij}, \sigma_y^2)\\ \alpha_j &\sim N(\mu_\alpha, \sigma^2_{\alpha})\\ \beta_j &\sim N(\mu_\beta, \sigma^2_{\beta}) \end{align}\]
Missing data and rstan
There are many packages in R that are capable of performing Bayesian inferences. Traditionally, people prefer using OpenBUG or JAGS, which mainly use Gibss sampler for obtaining samples. However, for the growing size of the data and complexity of the model structure, Gibss sampler may not be that efficient. People nowdays pay more attention on a more effictive sampling technique called Hamiltonian Monte Carlo (HMC), rstan are able to perform HMC, but the default sampler is known as “No-U-Turn Sampler’’”, according to Hoffman and Gelman (2014), it is a upgraded method of HMC which “Adaptively Setting Path Lengths in HMC”.
One advantage of JAGS over rstan is that JAGS can read NA value in input data and treat them as missing data automatically. But consider the performance between two different sampling methods, it may be worth exploring a little of handling missing data in rstan.
Example on CD4
In the following example, I will demonstrate fitting a LMM with missing data via rstan the data is obtaining from Dr. Patrick Breheny’s Bayesian Modeling in Biostatistics, according to the description, the data set contains repeated measurements on a sample of 251 children, each followed up for a period of one and a half years. In addition to CD4Pct, the data set also contains the following variables (I only include those I will use in the model):
\(\texttt{CD4Pct}\): The concentration CD4 Cells.
\(\texttt{ID}\): Each patient is assigned a unique ID. Note that there are several observations (rows) per patient.
\(\texttt{Visit}\): Ideally, patients were supposed to have visits at 1, 4, 7, 10, 13, 16, and 19 months, with 1 month being the initial visit. As is always the case in hospital data like this, the “16-month” visit did not take place exactly 15 months after the “1-month” visit, but Visit provides a rough approximation to the elapsed time between visits. Note that not all patients have an observation for each visit.
According to the EDA, we find that the \(\texttt{CD4Pct}\) itself does NOT follow a normal distrubtion but the square root transformation will be solving this problem, hence we will use \(Y_{ij} = \sqrt{\texttt{CD4Pct}_{ij}}\) be response.
First we read and manipulate the data
library(dplyr)
cd4 <- read.delim("data/cd4.txt", header = TRUE)
n_obs <- nrow(cd4)
J <- max(cd4$ID)
x <- seq(from = 1, by = 3, length.out = 7)
df <- tibble(id = rep(1:J, each = 7), visit = rep(x, J))
cd4 <- cd4 %>% select(id = ID, visit = Visit, CD4Pct) %>%
right_join(df, by = c('id', 'visit')) %>%
mutate(y = sqrt(CD4Pct))
n <- nrow(cd4)
cd4_data <- list(N = n, N_obs = n_obs, y_obs = cd4$y[1:n_obs], visit = cd4$visit, id = cd4$id, J = J)We consider a varying-intercept varying-slope model, we save the model in a stan file named cd4.stan, detail of the model is as follows,
data {
int<lower=0> N;
int<lower=0> N_obs;
int<lower=0> J;
vector[N_obs] y_obs;
int visit[N];
int<lower=1, upper=J> id[N];
}
transformed data{
int N_miss = N - N_obs;
}
parameters {
vector[N_miss] y_miss;
vector[J] alpha;
vector[J] beta;
real<lower=0> sigma_y;
real<lower=0> sigma_a;
real<lower=0> sigma_b;
real mu_a;
real mu_b;
}
transformed parameters{
vector[N] mu;
vector[N_obs] mu_obs;
vector[N_miss] mu_miss;
for(i in 1:N)
mu[i] = alpha[id[i]] + beta[id[i]] * visit[i];
mu_obs = head(mu, N_obs);
mu_miss = tail(mu, N_miss);
}
model {
mu_a ~ normal(0, 100);
mu_b ~ normal(0, 100);
sigma_y ~ uniform(0, 100);
sigma_a ~ uniform(0, 100);
sigma_b ~ uniform(0, 100);
for(j in 1:J){
alpha[j] ~ normal(mu_a, sigma_a);
beta[j] ~ normal(mu_b, sigma_b);
}
y_obs ~ normal(mu_obs, sigma_y);
y_miss ~ normal(mu_miss, sigma_y);
}
library(rstan)
# allow mutli core running at the same time to accerlate fitting process
options(mc.cores = 3)
fit <- stan(file='cd4.stan', data = cd4_data, chains = 3,
iter = 2500, warmup = 2000, refresh = 1000, save_warmup = FALSE)This will takes some time to compile the model and fitting MCMCs. This example takes about 45 second on my desktop. Once the fitting is done, we may extract the posterior samples and performing related analyses.
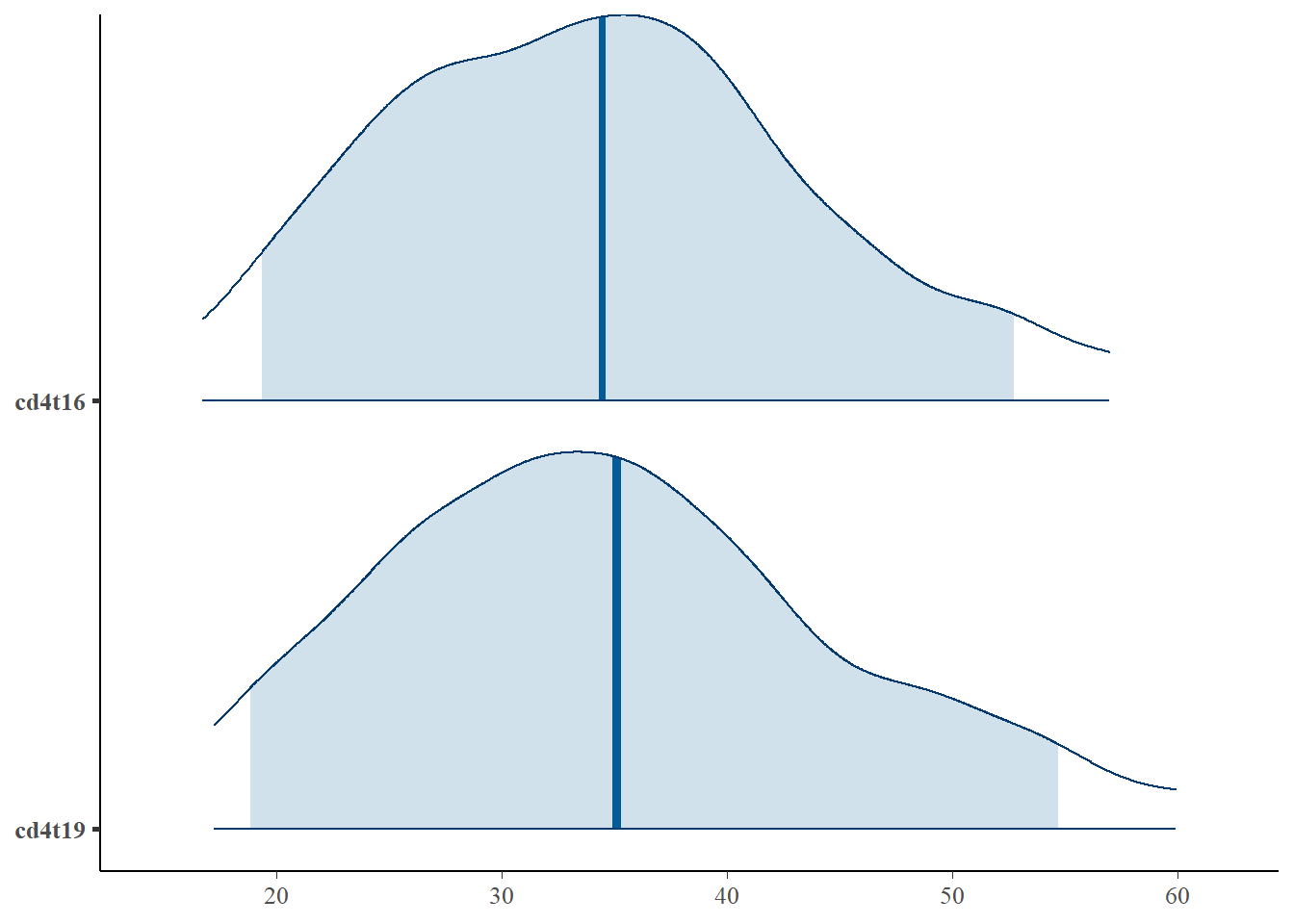
For example, we want to see the the prediction of missing response for ID = 18, also, we want to see the posterior distribution of the variance parameter \(\lbrace \sigma_y, \sigma_a, \sigma_b \rbrace\) by difference chains. The result can be plotted either by some built-in rstan function or a more flexible package called bayesplot.
idx <- which(cd4_data$id == 18)
idx[idx > cd4_data$N_obs] - cd4_data$N_obs
# position of y_miss for ID = 18
y18 <- extract(fit, pars = c('y_miss[38]', 'y_miss[39]'))
# permuted = FALSE, then will save data by chain
sigmas <- extract(fit, pars = c('sigma_y', 'sigma_a', 'sigma_b'), permuted = FALSE)
id18 <- tibble(t16 = post_param$`y_miss[38]`, t19 = post_param$`y_miss[39]`)
id18 <- id18 %>% mutate(cd4t16 = t16^2, cd4t19 = t19^2)library(bayesplot)
mcmc_areas(id18,
pars = c('cd4t16', 'cd4t19'),
prob = 0.9, # 90% intervals
prob_outer = 0.95, # 99%
point_est = "mean"
)
mcmc_dens_overlay(sigmas, pars = c('sigma_y', 'sigma_a', 'sigma_b'))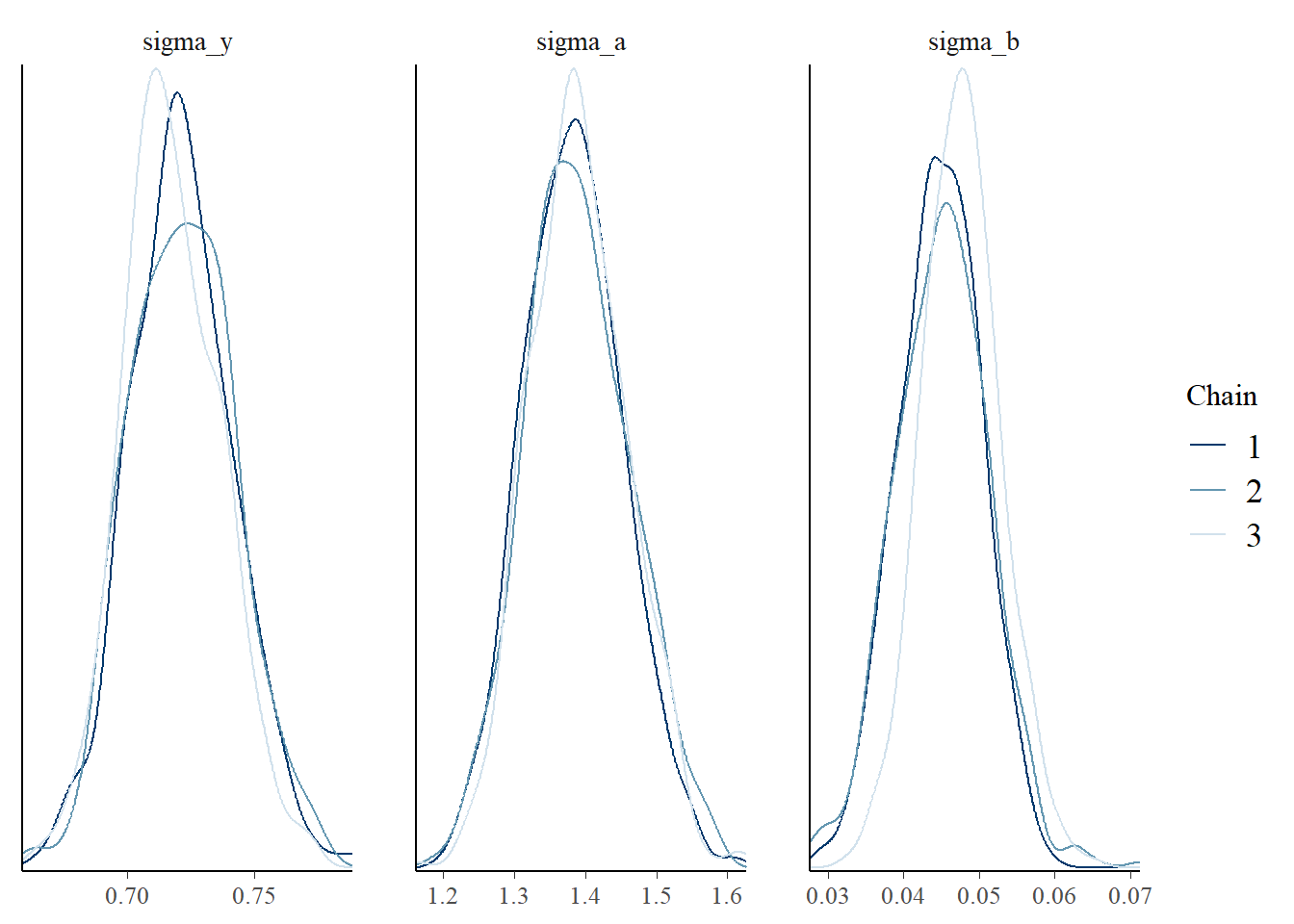
References:
Hoffman, M. D., & Gelman, A. (2014). The No-U-Turn sampler: adaptively setting path lengths in Hamiltonian Monte Carlo. J. Mach. Learn. Res., 15(1), 1593-1623.